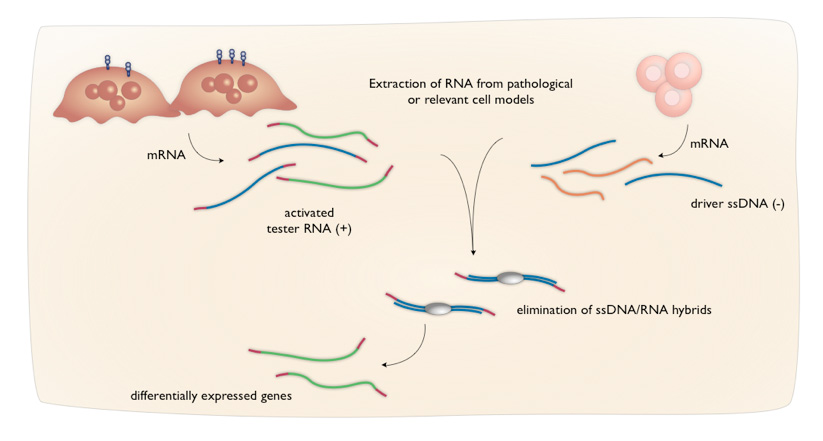
Subtractive Transcription-based Amplification of mRNA (STAR)
Alethia is in the business of discovering disease-related genes and validating their use as potential targets for the development of therapeutic products. Any changes in the condition of a cell, such as those occurring in a disease state or during drug treatment are caused by alterations in the activity of various genes in the cell, resulting in the change of expression of certain mRNAs. The differences in gene activity between two or more cells, as measured by the expressed mRNAs, is referred to as differential expression.
A typical human cell expresses approximately 20% to 30% of the genes found in the human genome as distinct mRNAs, of which nearly all (99%) are considered rare. Although each one of these rare mRNAs individually represent less than 0.005% of the total mRNAs, collectively they account for about half of the total mRNA in a typical cell. The functional and morphological differences between cell types are due to differential gene expression of different genes by cells with the same genome. Due to these complexities, determining the differentially expressed mRNA sequences between two or more cells can be a formidable task.
Alethia recognized a need for new technologies which reduce the complexity of cellular mRNA expression to a manageable number of differentially expressed sequences and generate simplified libraries of sequences directly usable in the validation process.
STAR Discovery Technology
In keeping with this mission, Alethia exploits a technology called Subtractive Transcription-based Amplification of mRNA (STAR) especially for the discovery of differentially expressed mRNA sequences including the rarest ones. The mRNA from each cellular sample is used for the preparation of specialized cDNA libraries, from which single-stranded (+) sense tester RNA and single-stranded (-) sense driver DNA can be generated. The tester RNA contains terminal sequences that allow amplification in an RNA amplification process. STAR preferentially amplifies those sequences uniquely present in the tester RNA that are absent or less abundant in the driver DNA through a combination of subtractive hybridization and RNA amplification.
What distinguishes the STAR technology is its capability to discover differentially expressed sequences in a completely opposite manner from that of previously developed methods. Unlike other subtractive methodologies, which require the reannealing of a double-stranded DNA for PCR-based amplification, STAR uses DNA from one library to inactivate single-stranded RNA from another library, leaving the remaining differentially expressed RNAs for linear amplification. It is this reversal of the subtractive hybridization paradigm that results in the preservation and subsequent detection of differentially expressed rare sequences.
STAR is distinct among all of the other commercial methods for differential expression analysis in its use of an RNA amplification process with linear kinetics, rather than DNA amplification with exponential kinetics. This minimizes the biased amplification of preferred sequences in a mixture that has been reported for techniques using PCR. Moreover, transcription-based amplification is recognized as the least biased method for preparing probes from mRNA for expression analysis on DNA arrays.
Since STAR is a preparative technology, it has distinct advantages over the various commercial versions of expression profiling, which are generally used for analytical purposes. The information on changes to the disease model resulting in differential expression is actually conveyed by the products of the STAR process. Thus, where other methods provide only expression data, STAR delivers informative material, infomaterial, which can be used directly for the validation of new therapeutic targets.